| verb | action |
|---|---|
| select() | choose columns by name |
| filter() | select rows based on conditions |
| arrange() | reorder the rows |
| summarise() | reduce raw data to user defined summaries |
| group_by() | group the rows by a specified column |
| mutate() | create a new variable |
6 Introduction to dplyr
6.1 Learning Objectives
By the end of this chapter, you will be able to:
Explain what the
dplyrWickham et al. (2023) package is and why it’s central to data wrangling.Use the six core verbs of dplyr (
select,filter,arrange,mutate,summarise, andgroup_by).Chain multiple operations together using the pipe operator (
|>).Apply
group_by()withsummarise()to generate grouped summaries.
6.2 dplyr
The dplyr package (part of the tidyverse Wickham (2023)) provides simple, consistent functions for cleaning and transforming data. These are the building blocks for almost every data workflow in R. Once you understand these verbs, you can read and write tidy R code fluently.
Try running the following functions directly in your consoleThe R console is the interactive interface within the R environment where users can type and execute R code. It is the place where you can directly enter commands, see their output, and interact with the R programming language in real-time.
6.3 Overview of Key Functions
6.4 Select
If we wanted to create a dataset that only includes certain variables, we can use the dplyr::select() function from the dplyr package.
For example I might wish to create a simplified dataset that only contains species, sex, flipper_length_mm and body_mass_g.
Run the below code to select only those columns
Alternatively you could tell R the columns you don’t want e.g.
Note that select() does not change the original penguins tibble. It spits out the new tibble directly into your console.
If you don’t save this new tibble, it won’t be stored. If you want to keep it, then you must create a new object.
6.5 Filter
Having previously used dplyr::select() to select certain variables, we will now use dplyr::filter() to select only certain rows or observations. For example only Adelie penguins.
We can do this with the equivalence operator ==
We can use several different operators to assess the way in which we should filter our data that work the same in tidyverse or base R.
| Operator | Name |
|---|---|
| A < B | less than |
| A <= B | less than or equal to |
| A > B | greater than |
| A >= B | greater than or equal to |
| A == B | equivalence |
| A != B | not equal |
| A %in% B | in |
If you wanted to select all the Penguin species except Adelies, you use ‘not equals’.
This is the same as
You can include multiple expressions within filter() and it will pull out only those rows that evaluate to TRUE for all of your conditions.
6.6 Arrange
The function arrange() sorts the rows in the table according to the columns supplied. For example
The data is now arranged in alphabetical order by sex. So all of the observations of female penguins are listed before males.
You can also reverse this with desc()
6.7 Mutate
Sometimes we need to create a new variable that doesn’t exist in our dataset. For example we might want to figure out what the flipper length is when factoring in body mass.
To create new variables we use the function mutate().
Note that as before, if you want to save your new column you must save it as an object. Here we are mutating a new column and attaching it to the new_penguins data oject.
6.8 Pipes
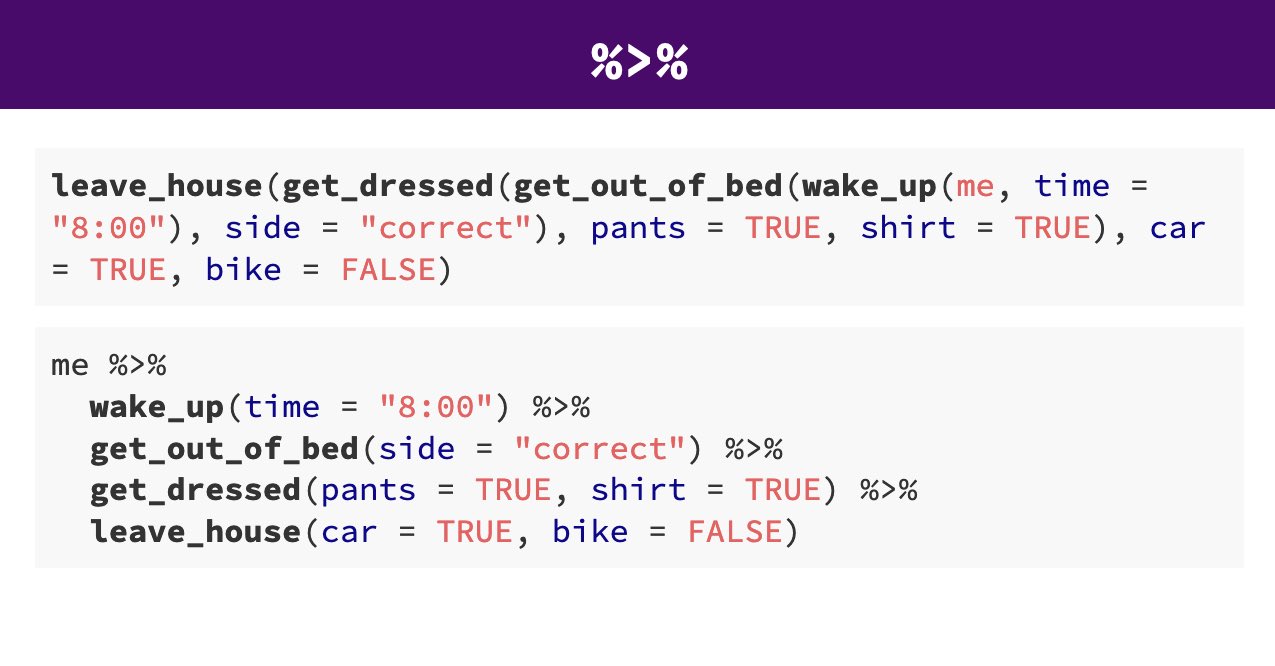
Pipes look like this: |> , a pipeAn operator that allows you to chain multiple functions together in a sequence. allows you to send the output from one function straight into another function. Specifically, they send the result of the function before |> to be the first argument of the function after |>. As usual, it’s easier to show, rather than tell so let’s look at an example.
The reason that this function is called a pipe is because it ‘pipes’ the data through to the next function. When you wrote the code previously, the first argument of each function was the dataset you wanted to work on. When you use pipes it will automatically take the data from the previous line of code so you don’t need to specify it again.
Your turn
Take any command you wrote earlier and rewrite it using pipes.
From R version 4 onwards there is now a “native pipe” |>
This doesn’t require the tidyverse magrittr package and the “old pipe” %>% or any other packages to load and use.
You may be familiar with the magrittr pipe or see it in other tutorials, and website usages. The native pipe works equivalntly in most situations but if you want to read about some of the operational differences, this site does a good job of explaining .
Your turn
Check your RStudio settings to see if you are using the Native Pipe
The Shortcut key is (Ctrl/Cmd) + Shift + M
6.9 Group by
The group_by() function is used to tell R that you want to perform operations within groups of your data rather than across the whole dataset.
Think of it as adding a “temporary label” to each row that says which group it belongs to. Then, when you use functions like summarise(), R performs the summary for each group separately.
Let’s say you want to find the average flipper length for each penguin species.
Without grouping we get a single value:
| mean_flipper |
|---|
| 200.9152 |
With grouping:
Gives one number per species
| Species | mean_flipper |
|---|---|
| Adelie Penguin (Pygoscelis adeliae) | 189.9536 |
| Chinstrap penguin (Pygoscelis antarctica) | 195.8235 |
| Gentoo penguin (Pygoscelis papua) | 217.1870 |
6.9.1 Grouping by multiple variables
| Species | Sex | mean_flipper |
|---|---|---|
| Adelie Penguin (Pygoscelis adeliae) | FEMALE | 187.7945 |
| Adelie Penguin (Pygoscelis adeliae) | MALE | 192.4110 |
| Adelie Penguin (Pygoscelis adeliae) | NA | 185.6000 |
| Chinstrap penguin (Pygoscelis antarctica) | FEMALE | 191.7353 |
| Chinstrap penguin (Pygoscelis antarctica) | MALE | 199.9118 |
| Gentoo penguin (Pygoscelis papua) | FEMALE | 212.7069 |
| Gentoo penguin (Pygoscelis papua) | MALE | 221.5410 |
| Gentoo penguin (Pygoscelis papua) | NA | 215.7500 |
You can think of group_by() as sorting your dataset into bins, one bin per group. Then, when you summarise() or mutate(), you’re performing that operation inside each bin.