19 Complex models
19.1 Designing a Model
We are introduced to the fruitfly dataset Partridge and Farquhar (1981)7. From our understanding of sexual selection and reproductive biology in fruit flies, we know there is a well established 'cost' to reproduction in terms of reduced longevity for female fruitflies. The data from this experiment is designed to test whether increased sexual activity affects the lifespan of male fruitflies.
The flies used were an outbred stock, sexual activity was manipulated by supplying males with either new virgin females each day, previously mated females ( Inseminated, so remating rates are lower), or provide no females at all (Control). All groups were otherwise treated identically.
type: type of female companion (virgin, inseminated, control(partners = 0))
longevity: lifespan in days
thorax: length of thorax in micrometres (a proxy for body size)
sleep: percentage of the day spent sleeping
19.2 Hypothesis
Before you start any formal analysis you should think clearly about the sensible parameters to test. In this example, we are most interested in the effect of sexual activity on longevity. But it is possible that other factors may also affect longevity and we should include these in our model as well, and we should think hard about what terms might reasonably be expected to interact with sexual activity to affect longevity.
19.3 Checking the data
You should now import, clean and tidy your data. Making sure it is in tidy format, all variables have useful names, and there are no mistakes, missing data or typos.
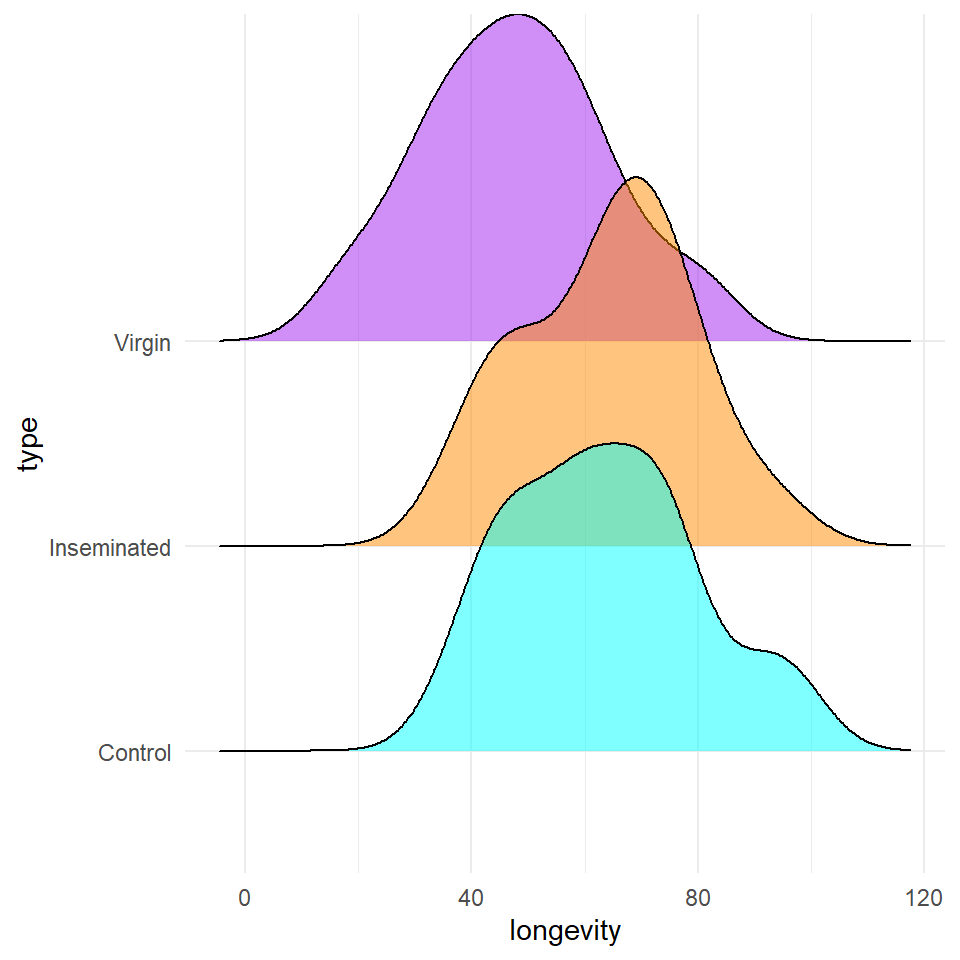
Based on the variables you have decided to test you should start with some simple visualisations, to understand the distribution of your data, and investigate visually the relationships you wish to test.
This is a full two-by-two plot of the entire dataset, but you should try and follow this up with some specific plots.
GGally::ggpairs(fruitfly)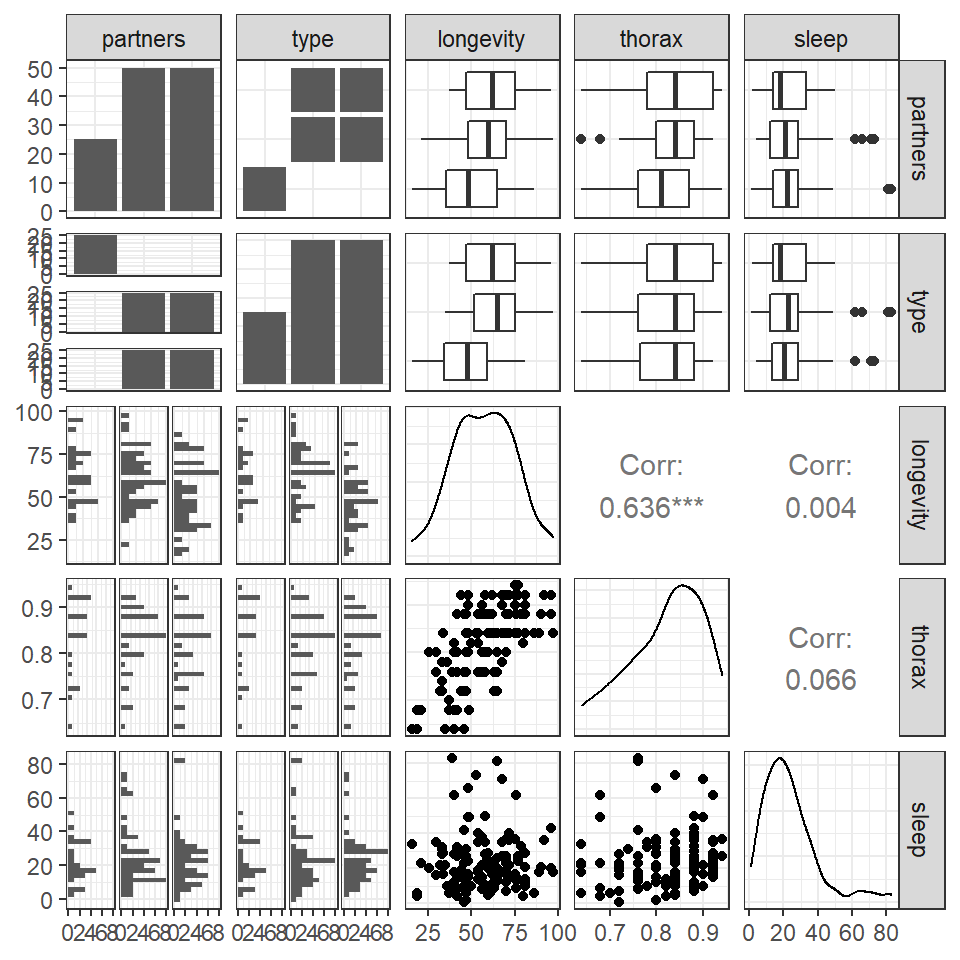
19.4 Activity 1: Think about your data
Think carefully about the plots you should make to investigate the potential differences and relationships you wish to investigate - try and answer the questions first before checking the examples hidden behind dropdowns.
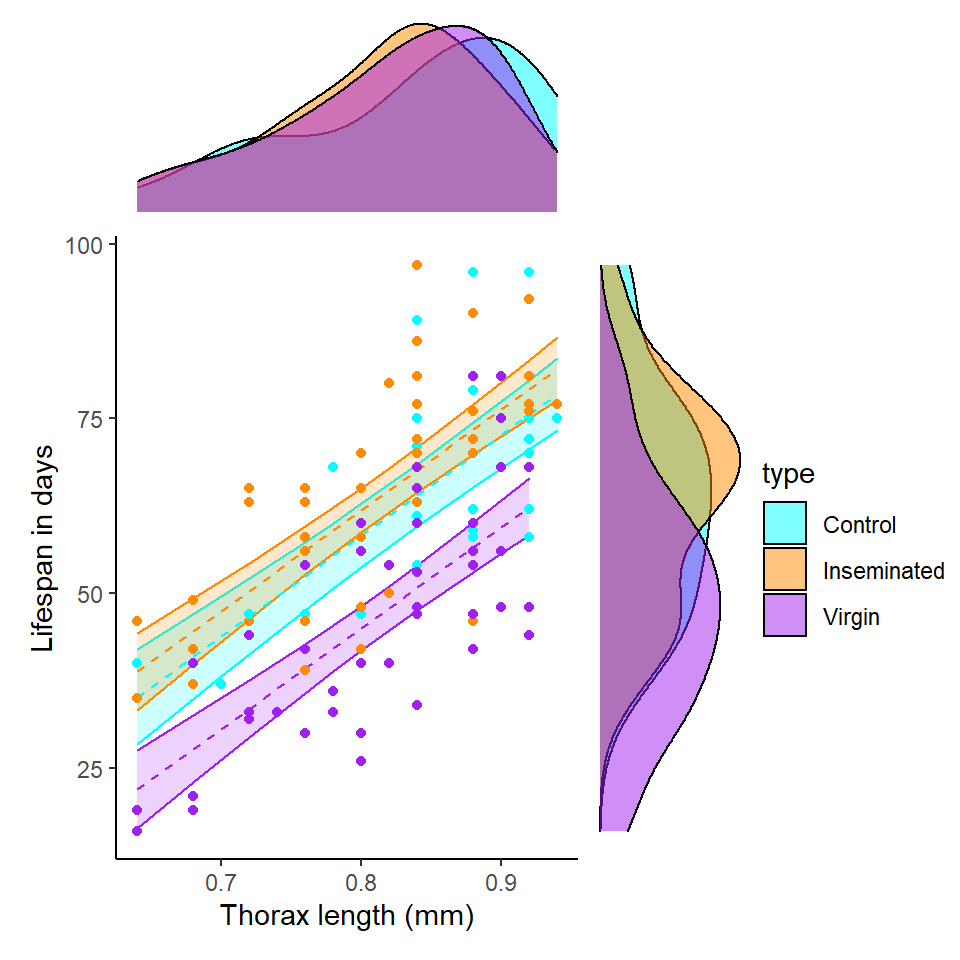
Q Does it like treatment affects longevity?
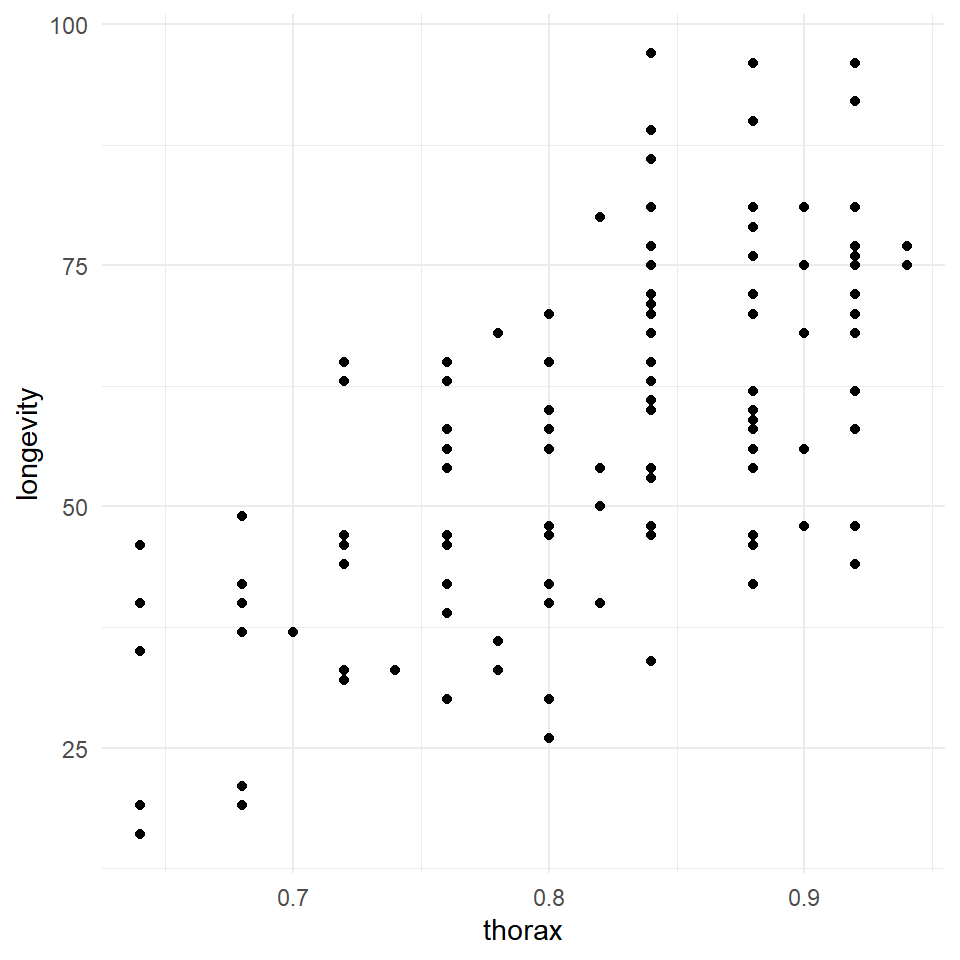
Q Does it look like size affects longevity?
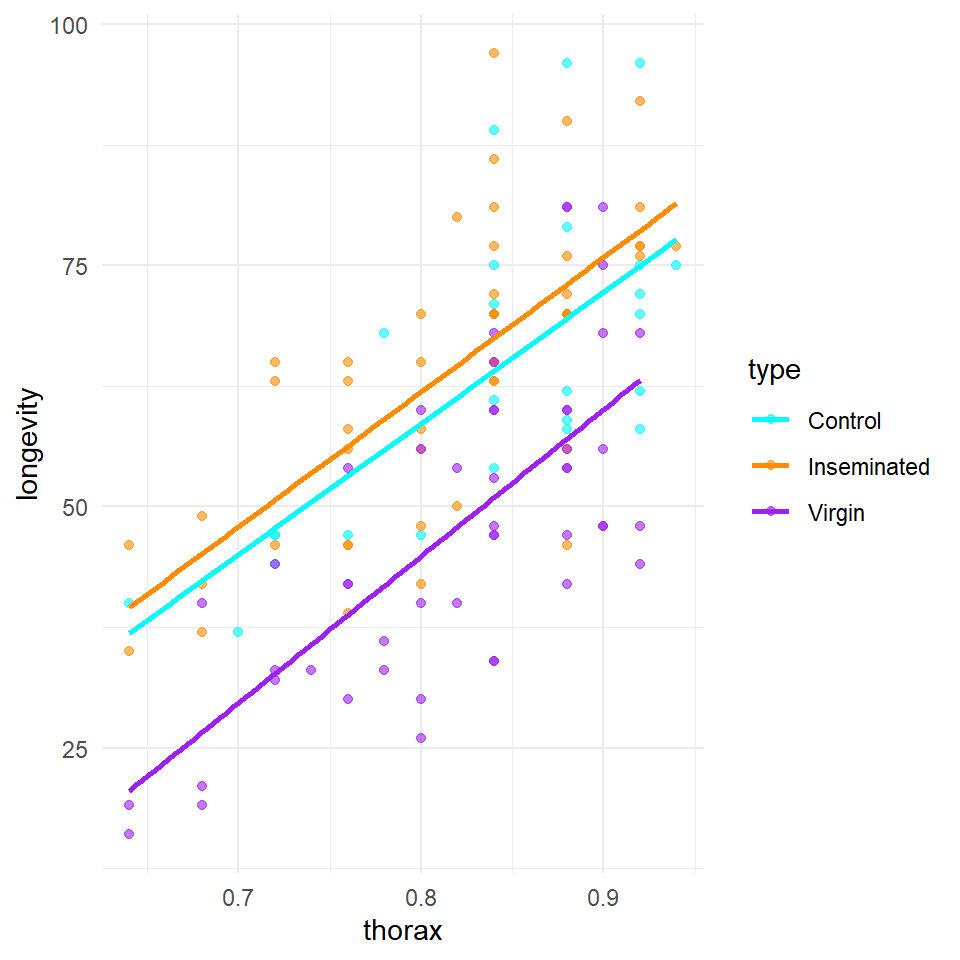
Q Does it look like size affects longevity differently between treatment groups?
Here it does look as though larger flies have a longer lifespan than smaller flies. But there appears to be little difference in the angle of the slopes between groups. This does not mean we can't test this in our model, but we may decide it is not worth including.
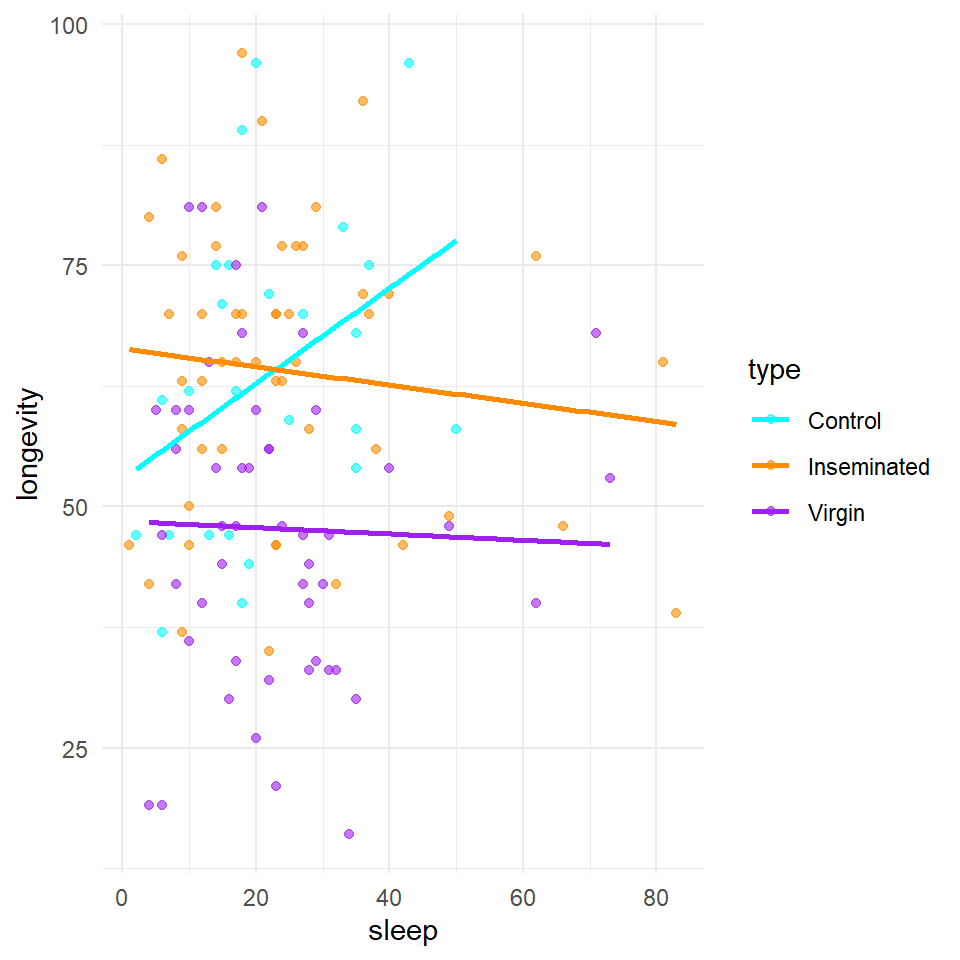
We are also interested in the potential effect of sleep on activity, we can construct a scatter plot of sleep against longevity, while including treatment as a covariate.
In these plots - Are the trendlines moving in the same direction?
Here it does look as though sleep interacts with treatment to affect lifespan. As the slopes of the lines are very different in each group. But in order to know the strength of this association, and if it is significantly different from what we might observe under the null hypothesis, we will have to build a model.
19.5 Designing a model
When you include an interaction term, the numbers produced from this are how much more or less the mean estimate is than if you just combined the main effects.
# a full model
flyls1 <- lm(longevity ~ type + thorax + sleep + type:sleep, data = fruitfly)
flyls1 %>%
broom::tidy()| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -57.5275383 | 11.3554560 | -5.0660703 | 0.0000015 |
| typeInseminated | 7.9883828 | 5.3412012 | 1.4956154 | 0.1374236 |
| typeVirgin | -10.9075381 | 5.4745755 | -1.9923989 | 0.0486358 |
| thorax | 142.5090010 | 13.4115350 | 10.6258531 | 0.0000000 |
| sleep | 0.0904459 | 0.1885893 | 0.4795919 | 0.6324053 |
| typeInseminated:sleep | -0.1965054 | 0.2082301 | -0.9436937 | 0.3472544 |
| typeVirgin:sleep | -0.1124276 | 0.2166543 | -0.5189260 | 0.6047842 |
Because we have included an interaction effect the number of terms is quite long and takes more consideration to understand. We can see for the individual estimates that it does not appear that the interaction is having a strong effect (estimate) and this does not appear to be different from a null hypothesis of no interaction effect. But we we should use an F test to look at the overall effect to be sure.
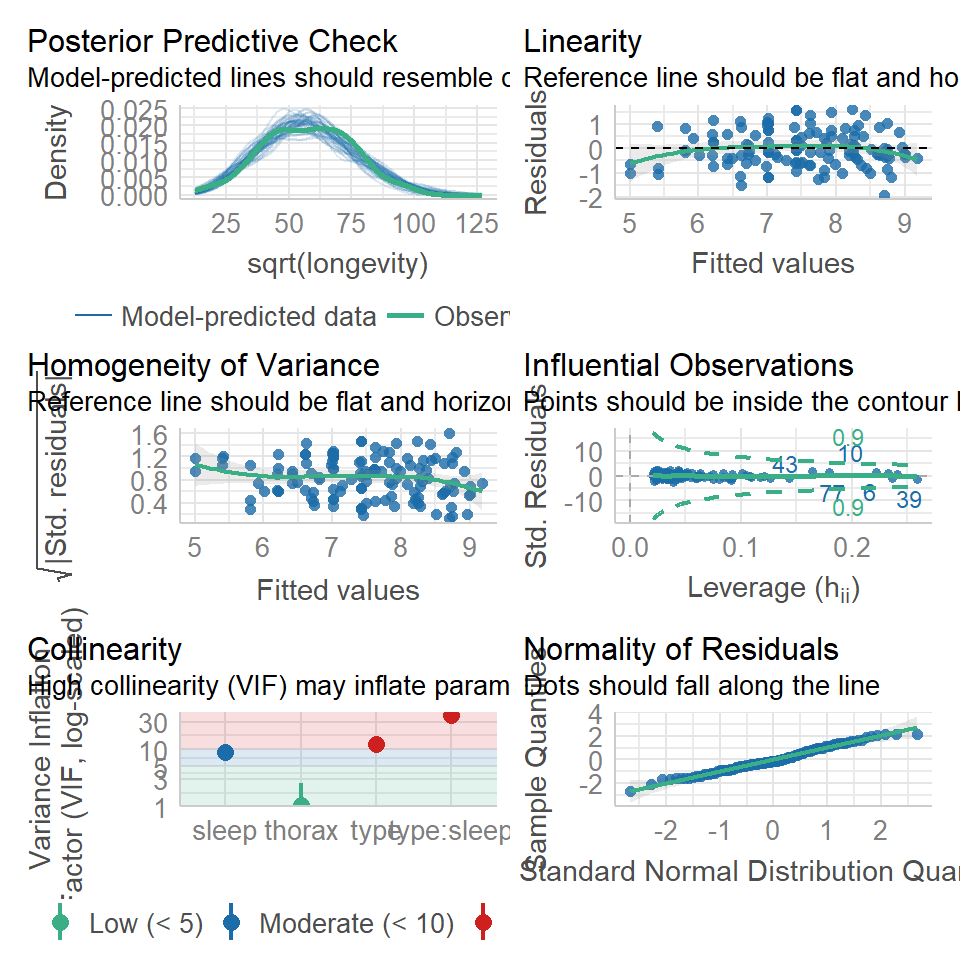
19.6 Model checking & collinearity
Before we start playing with the terms in our model, we should check to see if this is even a good way of fitting and measuring our data. We should check the assumptions of our model are being met.
performance::check_model(flyls1)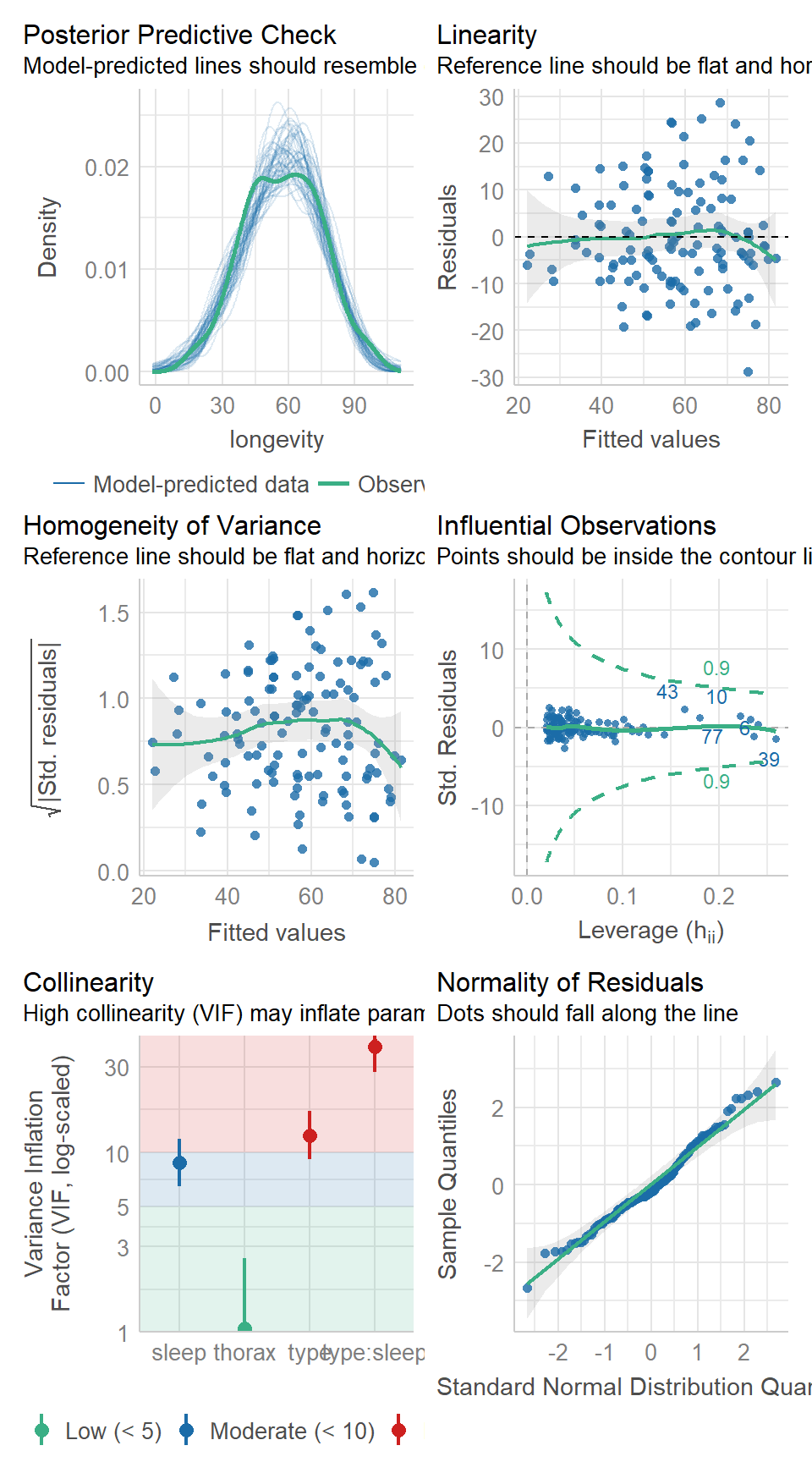
19.7 Activity 2: Model checking
Question - IS the assumption of homogeneity of variance met?
Question - ARE the residuals normally distributed?
Question - IS their an issue with Collinearity?
19.8 Data transformations
The most common issues when trying to fit simple linear regression models is that our response variable is not normal which violates our modelling assumption. There are two things we can do in this case:
-
Variable transformation e.g
lm(sqrt(x) ~ y, data = data)Can sometimes fix linearity
Can sometimes fix non-normality and heteroscedasticity (i.e non-constant variance)
Generalized Linear Models (GLMs) to change the error structure (i.e the assumption that residuals need to be normal - see next week.)
19.8.1 BoxCox
The BoxCox gets its name from its two inventors, George Box and David Cox. Implemented by the MASS package, when applied to a linear model it sytematically applies transformations by raising the y variable to a power (lambda).
The R output for the MASS::boxcox() function plots a maximum likelihood curve (with a 95% confidence interval - drops down as dotted lines) for the best transformation for fitting the data to the model.
| lambda value | transformation |
|---|---|
| 0.0 | log(Y) |
| 0.5 | sqrt(Y) |
| 1.0 | Y |
| 2.0 | Y^1 |
# run this, pick a transformation and retest the model fit
MASS::boxcox(flyls1)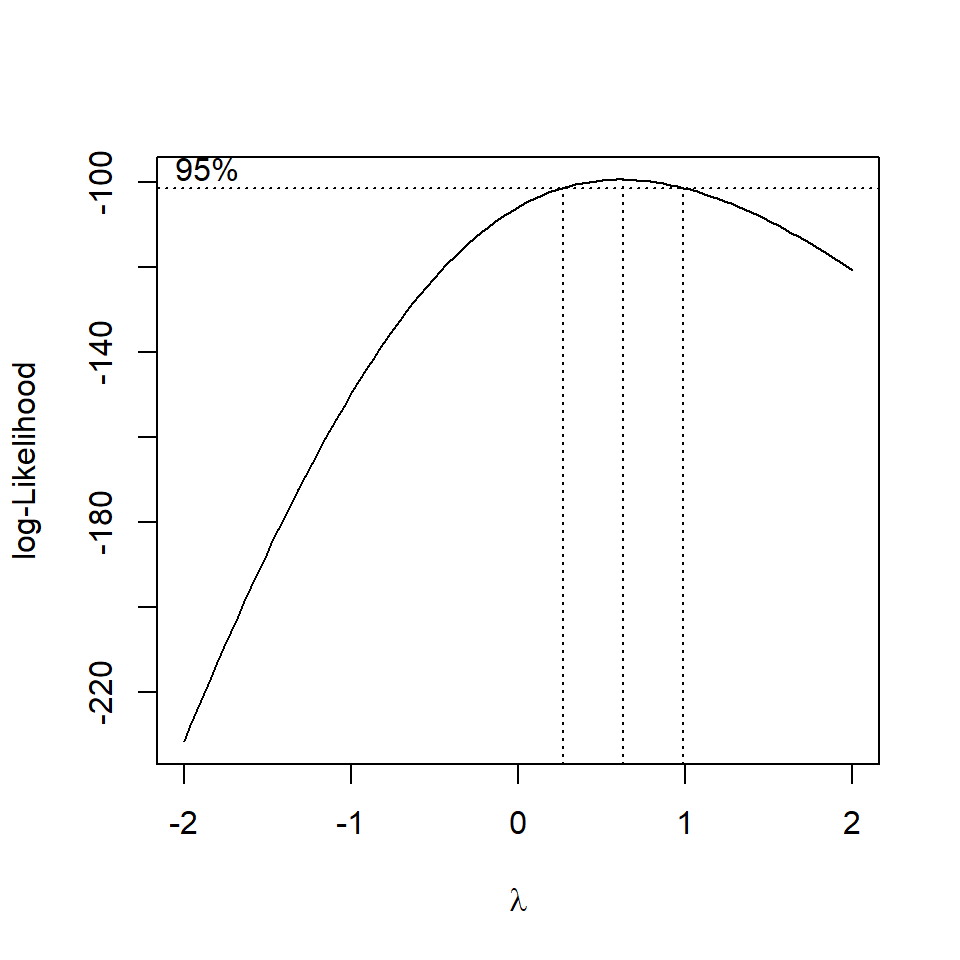
Figure 11.3: standard curve fitted by maximum likelihood, dashed lines represent the 95% confidence interval range for picking the 'best' transformation for the dependent variable
Question - Does the fit of the model improve with a square root transformation?
19.9 Model selection
# use drop1 function to remove top-level terms
drop1(flyls1, test = "F")| Df | Sum of Sq | RSS | AIC | F value | Pr(>F) | |
|---|---|---|---|---|---|---|
| <none> | NA | NA | 14994.43 | 612.3900 | NA | NA |
| thorax | 1 | 14347.4733 | 29341.90 | 694.3073 | 112.9087541 | 0.0000000 |
| type:sleep | 2 | 130.1431 | 15124.57 | 609.4702 | 0.5120865 | 0.6005695 |
Based on this ANOVA table, we do not appear to have a strong rationale for keeping the interaction term in the model (AIC or F-test). Therefore we can confidently remove the interaction, simplifying our model and making interpretation easier.
| Df | Sum of Sq | RSS | AIC | F value | Pr(>F) | |
|---|---|---|---|---|---|---|
| <none> | NA | NA | 15124.57 | 609.4702 | NA | NA |
| type | 2 | 7576.86233 | 22701.43 | 656.2337 | 30.057833 | 0.0000000 |
| thorax | 1 | 15282.82102 | 30407.39 | 694.7659 | 121.255596 | 0.0000000 |
| sleep | 1 | 86.27949 | 15210.85 | 608.1813 | 0.684551 | 0.4096663 |
Question - Should we drop sleep from this model?
19.10 Posthoc
Using the emmeans package is a very easy way to produce the estimate mean values (rather than mean differences) for different categories emmeans. If the term pairwise is included then it will also include post-hoc pairwise comparisons between all levels with a tukey test contrasts.
emmeans::emmeans(flyls2, specs = pairwise ~ type + thorax + sleep)## $emmeans
## type thorax sleep emmean SE df lower.CL upper.CL
## Control 0.821 23.5 61.3 2.26 120 56.8 65.8
## Inseminated 0.821 23.5 64.9 1.59 120 61.8 68.1
## Virgin 0.821 23.5 48.0 1.59 120 44.9 51.2
##
## Confidence level used: 0.95
##
## $contrasts
## contrast estimate SE df t.ratio
## Control 0.82096 23.464 - Inseminated 0.82096 23.464 -3.63 2.77 120 -1.309
## Control 0.82096 23.464 - Virgin 0.82096 23.464 13.25 2.76 120 4.796
## Inseminated 0.82096 23.464 - Virgin 0.82096 23.464 16.87 2.25 120 7.508
## p.value
## 0.3929
## <.0001
## <.0001
##
## P value adjustment: tukey method for comparing a family of 3 estimates
For continuous variables (sleep and thorax) - emmeans has set these to the mean value within the dataset, so comparisons are constant between categories at the average value of all continuous variables.
19.11 Activity 3: Write-up
19.12 Summary
In this chapter we have worked with our scientific knowledge to develop testable hypotheses and built statistical models to formally assess them. We now have a working pipeline for tackling complex datasets, developing insights and producing and explaining robust linear models.
19.12.1 Checklist
Think carefully about the hypotheses to test, use your scientific knowledge and background reading to support this
Import, clean and understand your dataset: use data visuals to investigate trends and determine if there is clear support for your hypotheses
Fit a linear model, including interaction terms with caution
Investigate the fit of your model, understand that parameters may never be perfect, but that classic patterns in residuals may indicate a poorly fitting model - sometimes this can be fixed with careful consideration of missing variables or through data transformation
Test the removal of any interaction terms from a model, look at AIC and significance tests
Make sure you understand the output of a model summary, sense check this against the graphs you have made
The direction and size of any effects are the priority - produce estimates and uncertainties. Make sure the observations are clear.
Write-up your significance test results, taking care to report not just significance (and all required parts of a significance test). Do you know what to report? Within a complex model - reporting t will indicate the slope of the line for that single term against the intercept, F is the overall effect of a predictor across all levels, post-hoc if you wish to compare across all levels.
Well described tables and figures can enhance your results sections - take the time to make sure these are informative and attractive.
19.13 Supplementary code
sjPlot A really nice package that helps produce model summaries for you automatically
| longevity | |||
|---|---|---|---|
| Predictors | Estimates | CI | p |
| (Intercept) | -56.05 | -78.18 – -33.91 | <0.001 |
| type [Inseminated] | 3.63 | -1.86 – 9.11 | 0.193 |
| type [Virgin] | -13.25 | -18.71 – -7.78 | <0.001 |
| thorax | 144.43 | 118.46 – 170.40 | <0.001 |
| sleep | -0.05 | -0.18 – 0.07 | 0.410 |
| Observations | 125 | ||
| R2 / R2 adjusted | 0.605 / 0.591 | ||
library(gtsummary)
tbl_regression(flyls2)| Characteristic | Beta | 95% CI1 | p-value |
|---|---|---|---|
| type | |||
| Control | — | — | |
| Inseminated | 3.6 | -1.9, 9.1 | 0.2 |
| Virgin | -13 | -19, -7.8 | <0.001 |
| thorax | 144 | 118, 170 | <0.001 |
| sleep | -0.05 | -0.18, 0.07 | 0.4 |
|
1
CI = Confidence Interval
|
|||